In previous posts I showed how to build a presence absence matrix (PAM) from species distribution data (check here) as PresenceAbsence object (check also how to transform a PAM into a PresenceAbsence). In this post I will show how to subset or crop a PresenceAbsence object.
Subseting a PresenceAbsence object can be done using the function lets.subsetPAM. You just need to provide the species you want to keep, and the PresenceAbsence object.
library(letsR)data("PAM")
set.seed(1)
names <- sample(PAM$Species_name, 20)
PAM_subset <- lets.subsetPAM(PAM, names)
par(mfrow = c(1, 2))
plot(PAM, main = "All species")
plot(PAM_subset, main = "Subset")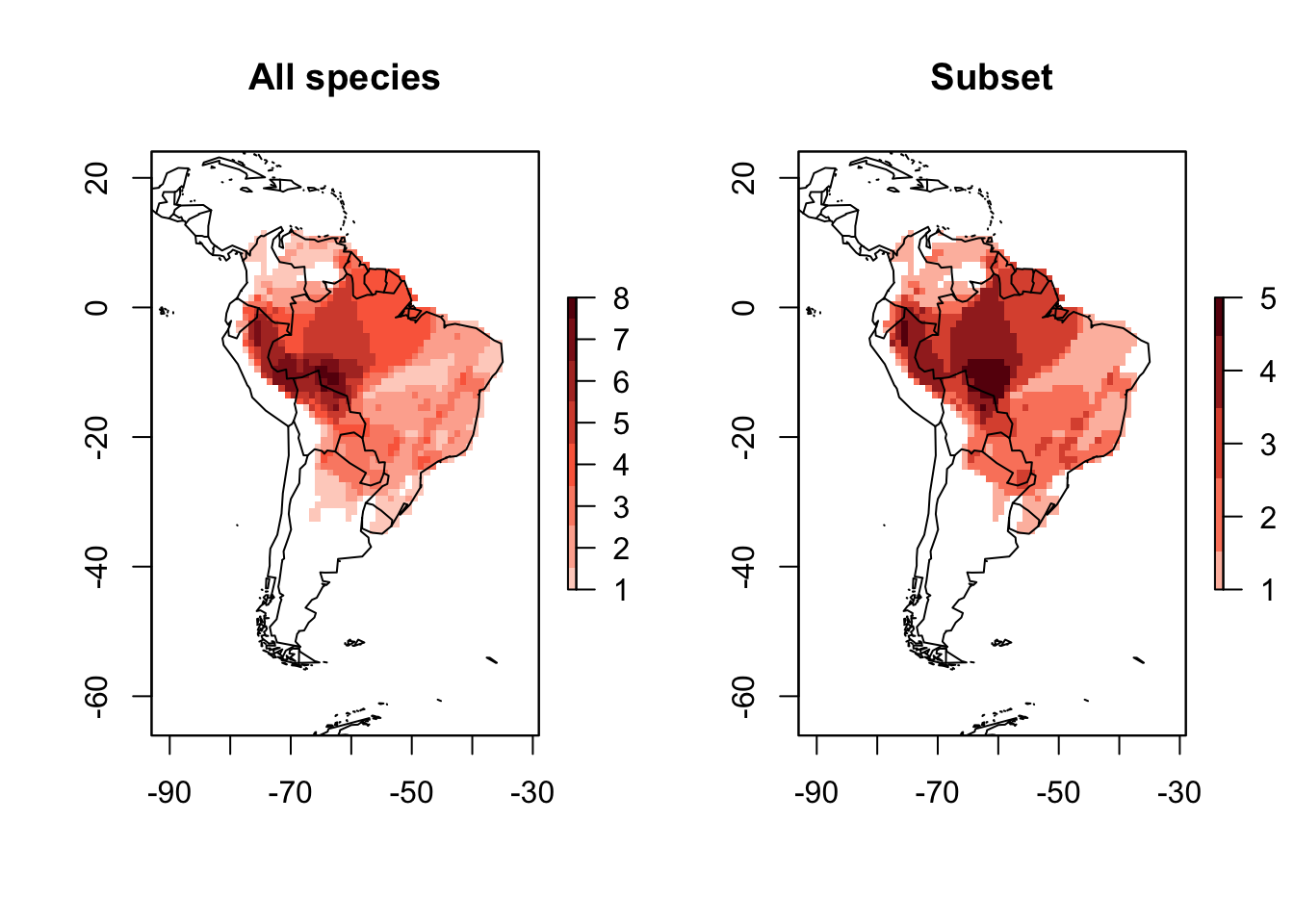
You may also want to crop the PresenceAbsence object to a determined region using a shapefile. The function lets.pamcrop is a easy way to do that. For example, lets crop our PAM to Brazil.
library(maptools) data(wrld_simpl) # World map
Brazil <- wrld_simpl[wrld_simpl$NAME == "Brazil", ] # Brazil (polygon)
PAM_crop <- lets.pamcrop(PAM, Brazil, remove.sp = TRUE)
par(mfrow = c(1, 2))
plot(PAM, main = "South America")
plot(PAM_crop, main = "Brazil",
col = colorRampPalette(c("darkgreen", "yellow", "blue")))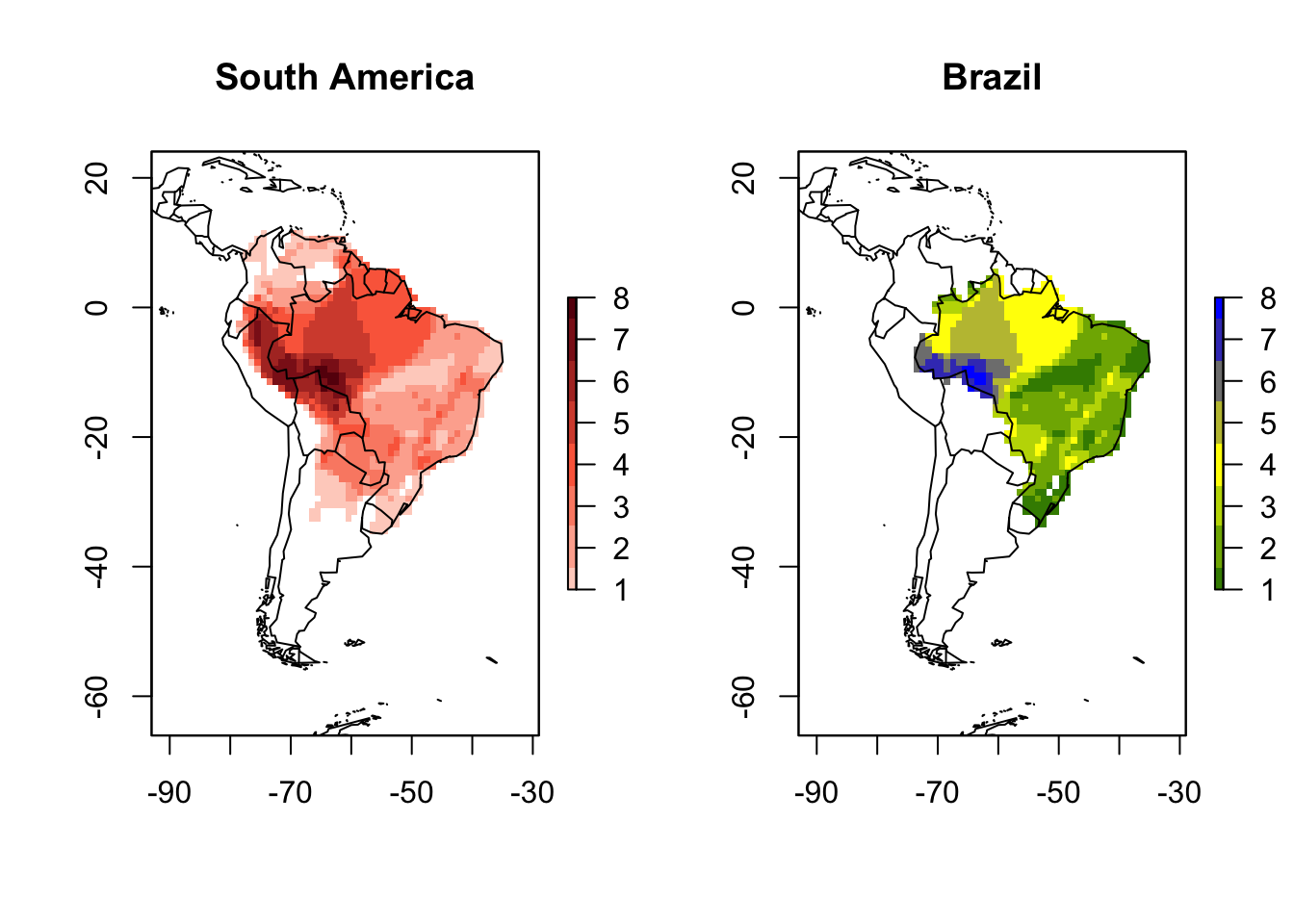
To cite letsR in publications use: Bruno Vilela and Fabricio Villalobos (2015). letsR: a new R package for data handling and analysis in macroecology. Methods in Ecology and Evolution. DOI: 10.1111/2041-210X.12401