Once you have transformed species distribution data into a presence absence matrix (PAM) in PresenceAbsence format (see the first and second posts on how to do it), you may want to add variables to it. These variables are normally in raster format, for example the WorldClim bioclimatic data, or in shapefile format, for example ecorregions of the world.
Adding variables in raster format
To add variables in raster format to a PresenceAbsence object we can use the function lets.addvar from the letsR package. This function takes a raster object with any resolution and extent, and transform it to match the information in your PresenceAbsence object. Once this is done, the variables are included as additional columns containing the aggregate/summarize value of the variable(s) in each cell of the presence-absence matrix. Let’s see an example using the bioclimatic data from WorldClim that can be downloaded with the package raster.
library(letsR)First we get the data.
r <- getData("worldclim", var = "bio",
res = 10)
plot(r)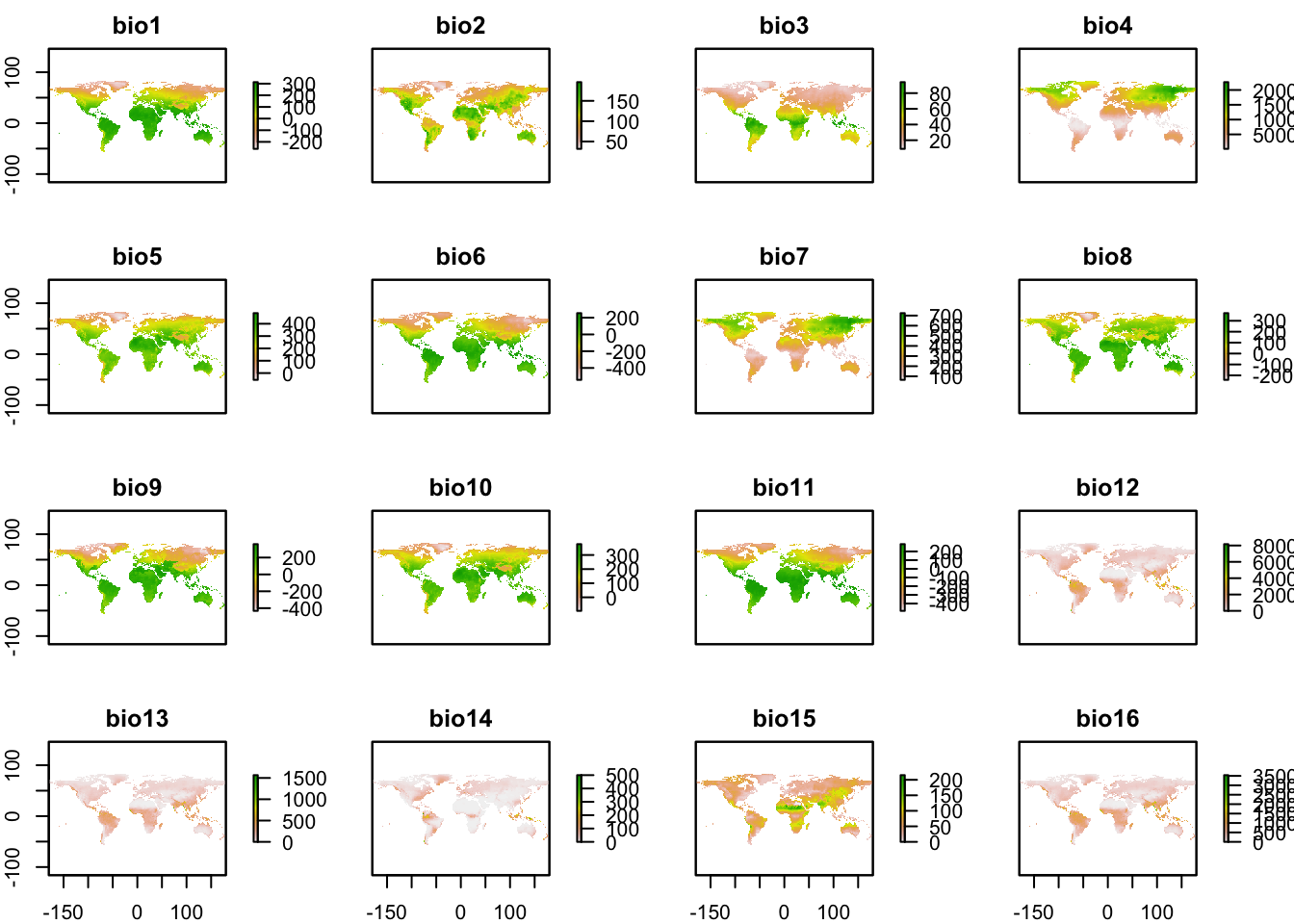
Here I will use the PresenceAbsence object for Phyllomedusa species generated previously.
data(PAM)
plot(PAM, main = "Phyllomedusa\nRichness")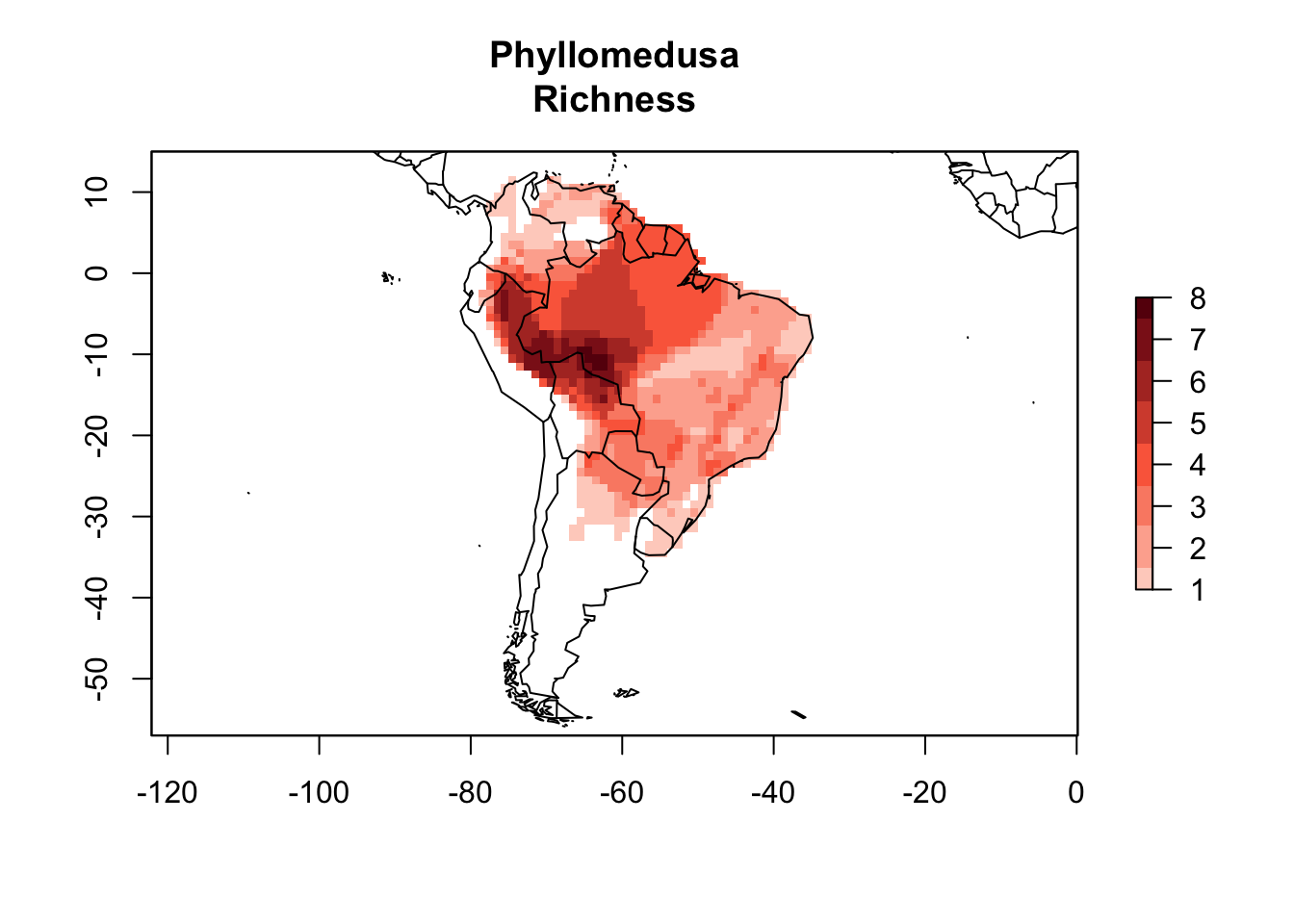
Let’s check the projection differences.
projection(r)## [1] "+proj=longlat +datum=WGS84 +ellps=WGS84 +towgs84=0,0,0"projection(PAM$Richness_Raster)## [1] "+proj=longlat +datum=WGS84"Since the CRS descriptions differ, we have to fix them first. In this case the projections are actually the same, but the character describing them differ, so we can just change the name directly. But if the projections are actually different, you may want to use the function raster::projectRaster.
projection(PAM$Richness_Raster) <- projection(r)We can now run the lets.addvar function.
PAM_env <- lets.addvar(PAM, r, fun = mean)The climatic data have a higher resolution than our PAM. In this case, we could choose a function to aggregate the values with the argument fun. In most of the situations, people will be interested in averaging values to aggregate multiple cells, but in some specific cases you may want to sum them, or get the standard deviation, or use any another function.
The result is a presence absence matrix, where the last columns now include the raster values. Check the table:
head(PAM_env)| Longitude(x) | Latitude(y) | Phyllomedusa araguari | Phyllomedusa atelopoides | Phyllomedusa ayeaye | Phyllomedusa azurea | Phyllomedusa bahiana | Phyllomedusa baltea | Phyllomedusa bicolor | Phyllomedusa boliviana | Phyllomedusa burmeisteri | Phyllomedusa camba | Phyllomedusa centralis | Phyllomedusa coelestis | Phyllomedusa distincta | Phyllomedusa duellmani | Phyllomedusa ecuatoriana | Phyllomedusa hypochondrialis | Phyllomedusa iheringii | Phyllomedusa itacolomi | Phyllomedusa megacephala | Phyllomedusa neildi | Phyllomedusa nordestina | Phyllomedusa oreades | Phyllomedusa palliata | Phyllomedusa perinesos | Phyllomedusa rohdei | Phyllomedusa sauvagii | Phyllomedusa tarsius | Phyllomedusa tetraploidea | Phyllomedusa tomopterna | Phyllomedusa trinitatis | Phyllomedusa vaillantii | Phyllomedusa venusta | bio1_mean | bio2_mean | bio3_mean | bio4_mean | bio5_mean | bio6_mean | bio7_mean | bio8_mean | bio9_mean | bio10_mean | bio11_mean | bio12_mean | bio13_mean | bio14_mean | bio15_mean | bio16_mean | bio17_mean | bio18_mean | bio19_mean |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -74.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 258.4000 | 91.65000 | 82.20000 | 526.7500 | 311.0500 | 200.1500 | 110.9000 | 258.7000 | 252.0000 | 263.5500 | 250.6000 | 1198.0500 | 250.7000 | 7.60000 | 80.50000 | 591.4000 | 34.15000 | 341.5500 | 108.0000 |
| -69.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 262.8497 | 84.22171 | 71.63269 | 892.6027 | 322.6034 | 206.4553 | 116.1482 | 262.5537 | 254.9960 | 272.4501 | 250.0026 | 731.0237 | 122.2032 | 17.98552 | 56.41115 | 309.3956 | 75.65198 | 198.3499 | 145.9311 |
| -68.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 266.2190 | 76.64375 | 73.65425 | 675.3595 | 319.1875 | 216.5294 | 102.6581 | 264.4078 | 261.5261 | 272.8083 | 256.0042 | 981.2149 | 166.8388 | 19.68305 | 57.18767 | 431.8224 | 98.00707 | 238.0230 | 229.3728 |
| -75.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 277.1437 | 90.83067 | 80.08133 | 426.1697 | 330.8697 | 217.9157 | 112.9540 | 276.3097 | 273.5883 | 281.3817 | 270.9663 | 1055.2210 | 205.3570 | 4.30900 | 73.66500 | 486.6277 | 19.99833 | 258.0923 | 109.8290 |
| -74.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 268.4548 | 108.15780 | 82.40520 | 445.8382 | 332.6014 | 202.0994 | 130.5020 | 266.5624 | 265.9968 | 273.0954 | 261.9868 | 1337.1702 | 237.9668 | 11.50960 | 66.55720 | 586.0000 | 52.64860 | 315.4482 | 273.1824 |
| -69.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 238.6514 | 100.90767 | 79.92085 | 588.9459 | 299.3219 | 173.6980 | 125.6239 | 239.7355 | 231.9007 | 244.1322 | 229.8309 | 808.9125 | 127.8143 | 12.74922 | 55.21132 | 338.7148 | 52.07358 | 233.9582 | 106.5961 |
If you do not want the coordinates and species included you can set the argument onlyvar = TRUE.
climate <- lets.addvar(PAM, r, fun = mean, onlyvar = TRUE)head(climate)| bio1_mean | bio2_mean | bio3_mean | bio4_mean | bio5_mean | bio6_mean | bio7_mean | bio8_mean | bio9_mean | bio10_mean | bio11_mean | bio12_mean | bio13_mean | bio14_mean | bio15_mean | bio16_mean | bio17_mean | bio18_mean | bio19_mean |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 258.4000 | 91.65000 | 82.20000 | 526.7500 | 311.0500 | 200.1500 | 110.9000 | 258.7000 | 252.0000 | 263.5500 | 250.6000 | 1198.0500 | 250.7000 | 7.60000 | 80.50000 | 591.4000 | 34.15000 | 341.5500 | 108.0000 |
| 262.8497 | 84.22171 | 71.63269 | 892.6027 | 322.6034 | 206.4553 | 116.1482 | 262.5537 | 254.9960 | 272.4501 | 250.0026 | 731.0237 | 122.2032 | 17.98552 | 56.41115 | 309.3956 | 75.65198 | 198.3499 | 145.9311 |
| 266.2190 | 76.64375 | 73.65425 | 675.3595 | 319.1875 | 216.5294 | 102.6581 | 264.4078 | 261.5261 | 272.8083 | 256.0042 | 981.2149 | 166.8388 | 19.68305 | 57.18767 | 431.8224 | 98.00707 | 238.0230 | 229.3728 |
| 277.1437 | 90.83067 | 80.08133 | 426.1697 | 330.8697 | 217.9157 | 112.9540 | 276.3097 | 273.5883 | 281.3817 | 270.9663 | 1055.2210 | 205.3570 | 4.30900 | 73.66500 | 486.6277 | 19.99833 | 258.0923 | 109.8290 |
| 268.4548 | 108.15780 | 82.40520 | 445.8382 | 332.6014 | 202.0994 | 130.5020 | 266.5624 | 265.9968 | 273.0954 | 261.9868 | 1337.1702 | 237.9668 | 11.50960 | 66.55720 | 586.0000 | 52.64860 | 315.4482 | 273.1824 |
| 238.6514 | 100.90767 | 79.92085 | 588.9459 | 299.3219 | 173.6980 | 125.6239 | 239.7355 | 231.9007 | 244.1322 | 229.8309 | 808.9125 | 127.8143 | 12.74922 | 55.21132 | 338.7148 | 52.07358 | 233.9582 | 106.5961 |
Now that we have the variables, we can use it to relate to our species data in many ways. For example, you could graph the relationship between precipitation/temperature and species richness.
library(ggplot2)rich <- rowSums(PAM$P[, -(1:2)])
mpg1 <- data.frame("Temperature" = climate[, 1]/10,
"Precipitation" = climate[, 12],
"Richness" = rich)
f1 <- ggplot(mpg1, aes(Temperature, Richness))
f1 <- f1 + geom_smooth(model = lm) +
geom_point(col = rgb(0, 0, 0, .6)) +
theme_bw()
f2 <- ggplot(mpg1, aes(Precipitation, Richness))
f2 <- f2 + geom_smooth(model = lm) +
geom_point(col = rgb(0, 0, 0, .6)) +
theme_bw()
f1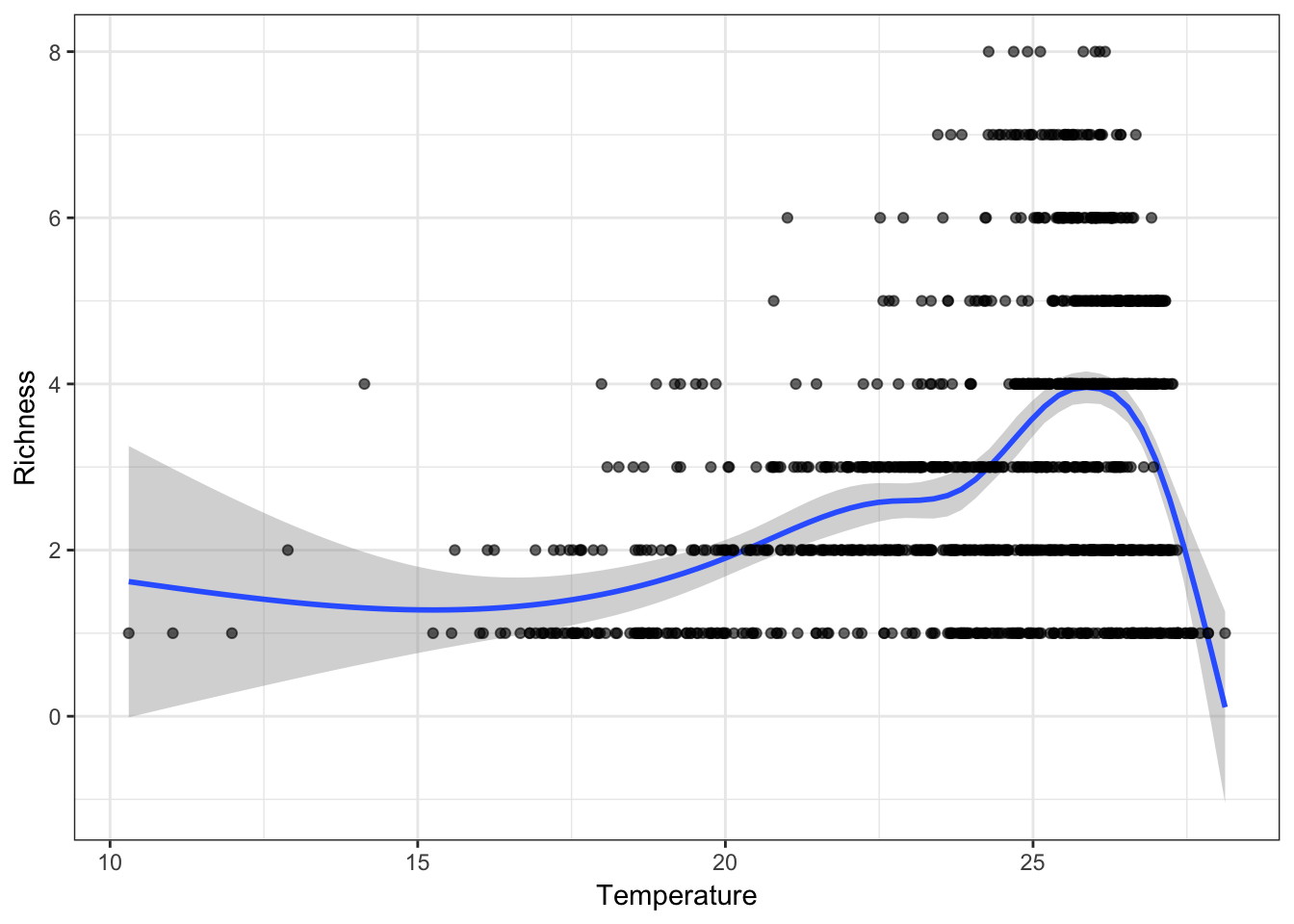
f2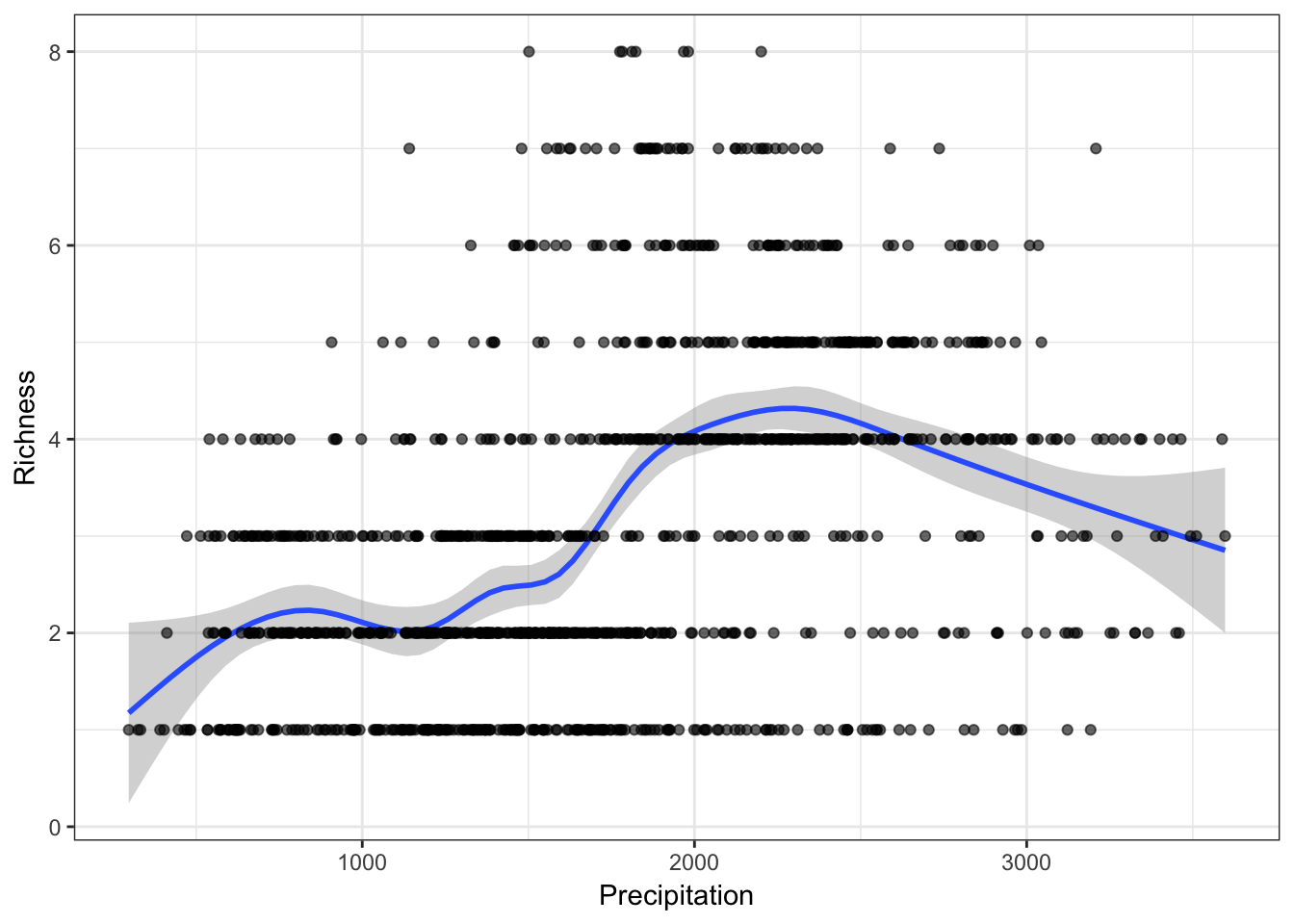
Adding variables in polygon format
Data in shapefile format like ecorregions, conservation units or countries, can be added to a PAM using the function lets.addpoly. This function adds polygons’ attributes as columns at the right-end of the matrix. The values represent the percentage of the cell covered by the polygon attribute used. As an example, we can use the South America countries map available in the package maptools.
library(maptools)data("wrld_simpl")
SA <- c("Brazil", "Colombia", "Argentina",
"Peru", "Venezuela", "Chile",
"Ecuador", "Bolivia", "Paraguay",
"Uruguay", "Guyana", "Suriname",
"French Guiana")
south_ame <- wrld_simpl[wrld_simpl@data$NAME %in% SA, ]
plot(south_ame)
Now we can add this variables to our PAM.
PAM_pol <- lets.addpoly(PAM, south_ame, "NAME")head(PAM_pol)| Longitude(x) | Latitude(y) | Phyllomedusa araguari | Phyllomedusa atelopoides | Phyllomedusa ayeaye | Phyllomedusa azurea | Phyllomedusa bahiana | Phyllomedusa baltea | Phyllomedusa bicolor | Phyllomedusa boliviana | Phyllomedusa burmeisteri | Phyllomedusa camba | Phyllomedusa centralis | Phyllomedusa coelestis | Phyllomedusa distincta | Phyllomedusa duellmani | Phyllomedusa ecuatoriana | Phyllomedusa hypochondrialis | Phyllomedusa iheringii | Phyllomedusa itacolomi | Phyllomedusa megacephala | Phyllomedusa neildi | Phyllomedusa nordestina | Phyllomedusa oreades | Phyllomedusa palliata | Phyllomedusa perinesos | Phyllomedusa rohdei | Phyllomedusa sauvagii | Phyllomedusa tarsius | Phyllomedusa tetraploidea | Phyllomedusa tomopterna | Phyllomedusa trinitatis | Phyllomedusa vaillantii | Phyllomedusa venusta | Argentina | Bolivia | Brazil | Chile | Colombia | Ecuador | French Guiana | Guyana | Suriname | Paraguay | Peru | Uruguay | Venezuela |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -74.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0.12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 |
| -69.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.58 |
| -68.5 | 11.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 |
| -75.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 |
| -74.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0.97 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 |
| -69.5 | 10.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.00 |
This information can be used to calculate the number of species per country for example.
vars_col <- (ncol(PAM$P) + 1):ncol(PAM_pol)
n <- length(vars_col)
rich_count <- numeric(n)
for (i in 1:n) {
rich_count[i] <- sum(colSums(PAM$P[PAM_pol[, vars_col[i]] > 0,
-(1:2)]) > 0)
}
labs <- as.factor(colnames(PAM_pol)[vars_col])
names(rich_count) <- labsmpg <- data.frame("Richness" = rich_count, "Country" = as.factor(labs))
g <- ggplot(mpg, aes(labs, Richness))
g + geom_bar(stat = "identity") + labs(x = "") +
theme(axis.text.x = element_text(angle = 90, hjust = 1))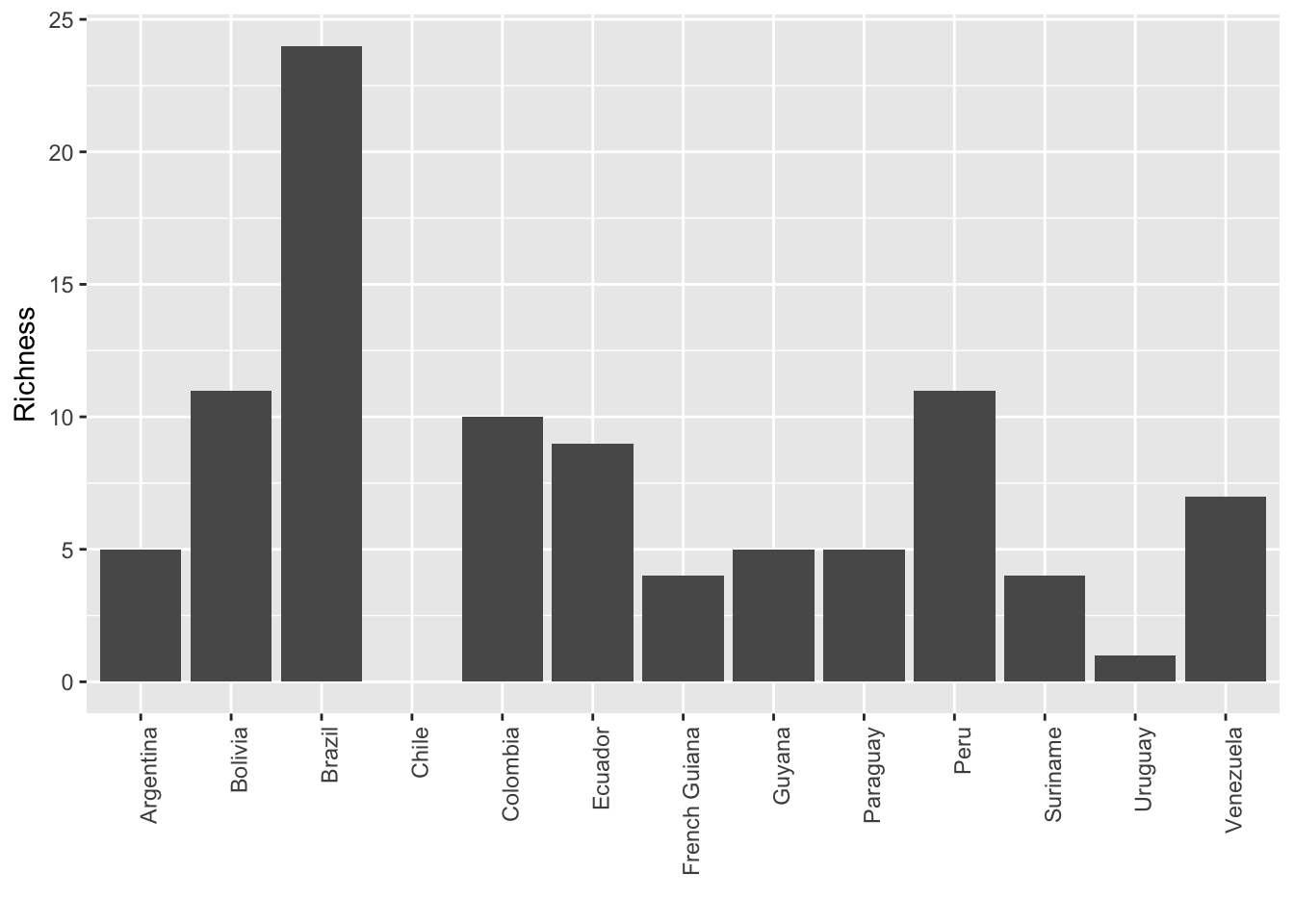
To cite letsR in publications use: Bruno Vilela and Fabricio Villalobos (2015). letsR: a new R package for data handling and analysis in macroecology. Methods in Ecology and Evolution. DOI: 10.1111/2041-210X.12401